< Previous lab day | Module 1 lab index | Next lab day >
Introduction
So far you have prepared the DNA encoding both the 6-5 and the 8-12 aptamer fragments. However, it is the secondary structure of the RNA that actually allows the 8-12 sequence to bind to heme. In order to make RNA from DNA, you will perform an in vitro transcription (IVT) reaction.
What is needed to create RNA from DNA? In PCR, you used a heat-stable DNA polymerase and dNTPs (deoxynucleotide triphosphates) to make your DNA. In an IVT, you will use an RNA polymerase and NTPs (nucleotide triphosphates) instead. The polymerase is derived from the T7 bacteriophage, and requires that the DNA to be copied contains the T7 promoter sequence, or TAA TAC GAC TCA CTA TAG GG. The buffer conditions are also somewhat different (check out the reagent lists at the end of each day), but both contain the important co-factor Mg2+. Finally, an IVT contains pyrophosphatase, because it has been empirically found to increase efficiency. Pyrophosphate is produced during the IVT, so by Le Chatelier's principle, reaction completion may be improved by removing this product.
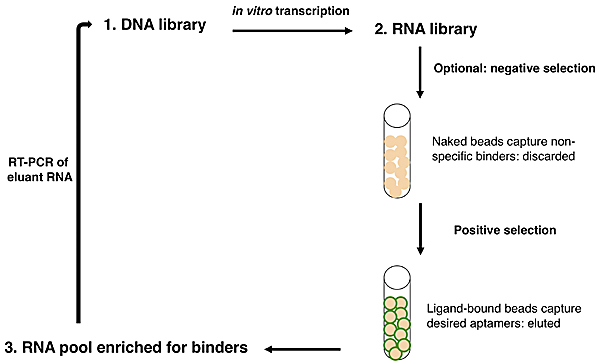
Overview of SELEX.
Now let's consider how an IVT fits into the overall scheme of SELEX, or systematic evolution of ligands by exponential enrichment. SELEX was simultaneously reported by two groups in 1990:
Ellington and Szostak described RNA that bound to select aromatic small molecules, while Tuerk and Gold isolated RNA that bound to a DNA polymerase. As shown in the figure at right, due to the stability of DNA one usually starts with a DNA library rather than directly with an RNA library. The DNA is copied into RNA by transcription, and the resulting RNA pool is run over an affinity column that has the ligand of interest bound to it. Aptamers that do not bind are washed away by flowing buffer through the column. Afterward, RNA that does bind to the ligand is washed off, either by using free ligand as a competitor or sometimes by other changes in buffer conditions (e.g., salt concentration or pH). This RNA pool is copied into DNA by reverse transcription, then amplified by PCR, often using a one-step kit these days. After RT-PCR, the new library (presumably containing many fewer species of DNA than the original library) is again transcribed to RNA. Now the new library may be further refined by a second column selection.
An optional but often important step is to perform a negative selection. To rid the RNA pool of aptamers that bind non-specifically to the column material, one can incubate the library with a column resin that lacks ligand but is otherwise identical to the affinity resin. After all, it would be a shame to think one has found several heme-binding aptamers, only to discover that they in fact bind to agarose beads! The supernatant from the negative selection is then used directly in the positive selection. Sometimes, multiple negative selections may be appropriate. For example, if one is interested in finding an aptamer that binds to a particular ligand but not to several similar ligands, one could perform negative selections using affinity columns presenting those those undesired ligands. No matter how many precautions one takes, the statistics of large numbers suggests that there will always be some false positives: RNA sequences that manage to get detectably amplified, but do not bind to the ligand of interest.
After one or more column selections, one may use cloning to select individual RNA species for sequencing or for testing in a functional assay. Again, ligating the DNA library into a plasmid that can be expressed in bacteria is easier than working with the RNA. DNA can be isolated from individual colonies of bacteria that each contain a single aptamer-encoding DNA, then sequenced and transcribed to RNA if desired. A typical starting library might have 10^13 to 10^15 sequences! In Szostak's early binding experiments, molecules that bound to the aromatic dyes were enriched from less than 1% to greater than 50% of the RNA library population after four rounds of selection, and the unique population was reduced to ~100-100,000 sequences. In our case, we already know the sequences of the two aptamers in question, and are simply interested in their ratio after a round of column selection. The functional assay that can indicate this ratio measures aptamer binding to heme by spectrophotometry.
The IVT will run during the whole lab period. In the meantime, we will discuss a journal article, both to learn more about RNA aptamers and to become comfortable reading and discussing the primary scientific literature. In 2-3 weeks, you will each present an article on your own.
Protocols
Because you are preparing RNA, you will have to take special precautions today and for the rest of the module. RNA is strikingly different from DNA in its stability. Consequently it is more difficult to work with RNA in the lab. It is not the techniques themselves that are difficult; indeed, many of the manipulations are nearly identical to those used for DNA. However, RNA is rapidly and easily degraded by RNases that exist everywhere. There are several rules for working with RNA. They will improve your chances of success. Please follow them all.
- Use warm water on a paper towel to wash lab equipment, like microfuges, before you begin your experiment. Then wipe them down with "RNase-away" solution.
- Wear gloves when you are touching anything that will touch your RNA.
- Change your gloves often.
- Before you begin your experiment clean your work area, removing all clutter. Wipe down the benchtop with warm water then "RNase-away," and then lay down a fresh piece of benchpaper.
- Use RNA-dedicated solutions and if possible RNA-dedicated pipetmen.
- Get a new box of pipet tips from the RNA materials area and label their lid "RNase FREE" if the lid is not yet labeled.
Part 1: In Vitro Transcription
The table below lists the amount of each reaction component needed for an 80 μL IVT. First, you should calculate how much total IVT Master Mix to make (2 rxns plus 10% excess) and check your calculations with the teaching faculty if desired. Next, you can aliquot the appropriate amount of Master Mix into a number of eppendorf tubes, then add the relevant DNA to each labeled tube.
| REAGENT | AMOUNT for 1 REACTION (μL) | AMOUNT for 2 REACTIONS + 10% |
|---|---|---|
| G7 buffer (2.5X stock) | 32 | |
| 1N KOH | 4.48 | |
| Pyrophosphatase | 4 | |
| NTPs | 22.4 | |
| T7 polymerase | 4 | |
| DNA | 13.1 | N/A |
Place your reaction tubes on the 37 °C heat block and write the time in your notebook as well as on the sheet at the front bench. After 4 hours, the reactions will be frozen at – 20 °C until next time. When everyone is ready, we will begin the journal article discussion.
Part 2: Choose Column Conditions
Before leaving today, you and your partner should sign up for one of the column selection protocols listed in the following table:
| PROTOCOL # | STUDENT PAIR | 8-12% | WASH # (PARAMETER 1) | WASH # (PARAMETER 2) | NOTE CHANGES TO PLAN HERE |
|---|---|---|---|---|---|
| 1 | 2 | 4 | 24 | ||
| 2 | 2 | 8 | 16 | ||
| 3 | 10 | 4 | 24 | ||
| 4 | 10 | 8 | 16 | ||
| 5 | 50 | 4 | 24 | ||
| 6 | 50 | 8 | 16 | ||
| 7 | 50 | 4 | 24 |
Journal Article Discussion
We will discuss this paper:
- Rusconi, C. P., et al. "Antidote-Mediated Control of an Anticoagulant Aptamer in vivo." Nature Biotechnology 22, no. 11 (November 2004): 1423-8.
Scientific papers are dense and often time-consuming to read and understand, but with practice, you will find strategies that improve your comprehension efficiency. Here's one tip to get you started: when reading newly reported results, be sure to refer to the associated figures frequently, because visual information is often easier to take in than purely verbal descriptions.
Technical Background
Several terms and assays in the Rusconi, et al. paper may be unfamiliar to you. (On that note, "assay" is just a term that means a type of measurement. For example, a cell viability assay measures the number of living cells in a sample.) Here we will provide some background on selected topics.
- Bolus refers to a one-time injection of drug, as opposed to a continuous infusion (via an IV line).
- Pharmacokinetics is the study of the route a drug takes in the body, both where it circulates to and how it is eliminated.
- Pharmacodynamics is the study of the actual effects, both intended and unintended, of the drug on the recipient, and of the drug's mechanism and molecular-level kinetics.
- Plasma is blood with the cells removed. (Serum is blood with both cells and clotting factors absent.)
Note that multiple pathways for blood clotting exist. An APTT assay measures the activity of the pathway that is dependent on the presence of clotting Factor IX. In Rusconi's work, buffer with or without aptamer was mixed with a plasma test sample, then activated with phospholipids and calcium ions. Adding an anti-coagulant should increase the time required for a clot to form in the activated mixture. The PT assay, on the other hand, measures the activity of a Factor IX-independent pathway. In this case, thromboplastin and calcium ions are used for plasma activation.
Discussion Topics
Writing
As you read the paper by Rusconi, et al., consider not only its scientific content, but also the authors' writing style (perhaps not all on one read!). Refer to our Guidelines for Writing Up Your Research Sketch out answers to the questions below (right on the paper if you wish). Your answers will not be collected, but you may be called on in discussion to share your ideas.
- Which part of the text corresponds with an Introduction section? What is the topic and/or function of each paragraph? What purpose(s) do the citations serve?
- Which part of the text corresponds with a Results section? Can you find one or more examples of paragraphs with effective introductory and concluding sentences, according to our Guidelines for results? Are there any parts in the Results that you think belong in the Discussion instead, according to our Guidelines for discussions and results vs. discussions?
- Which part of the text corresponds with a Discussion section? What is the topic and/or function of each paragraph? What purpose(s) do the citations serve?
Content
When you arrive in lab today, each group will be assigned one of the following topics to present to and discuss with the rest of the class. You should be somewhat familiar with the whole Rusconi, et al. paper by now, but will have some time in-class to refresh your memory and become the resident expert in one of the following areas.
- Introduction and Figure 1a
- What are the advantages of using an aptamer as an anti-coagulant drug?
- What related work have the authors previously done, and what research gap is being addressed by this new paper?
- Sumarize the sample and control aptamers used in this study. What is the aptamer target?
- Figure 1, b-e
- Describe the major findings portrayed in this figure.
- What was the purpose of testing the aptamer against multiple animal sources?
- Figure 2, a-c
- Describe the major findings portrayed in this figure.
- Why did the authors do these experiments, if they already had the data in Figure 1c?
- Figure 2, d-f
- Describe the major findings portrayed in this figure.
- How did the authors distinguish between two hypotheses about the antidote's potent action?
- Figure 3
- Describe the major findings portrayed in this figure.
- Briefly, what is a thrombus, and under what conditions is one likely to form? (May require research outside the paper.)
- What precautions did the authors take to avoid bias during data collection (see Methods)?
- Figure 4
- Describe the major findings portrayed in this figure.
- Briefly, what is a P-value? (May require research outside the paper.)
- What does the "vehicle" refer to (see Methods) and what is its role?
- Wrap-up
- How does the aptamer's potential as a drug compare to the current state-of-the-art?
- Briefly, search PubMed for whether Rusconi, et al. have published any further studies on the aptamers descrbied here, read the abstract(s), and summarize any progress.
For Next Time
-
Day 4 of this module is poised to run long, so you should read the entire protocol and perhaps prepare some of your lab notebook in advance. In addition, to save time later you should prepare an automated worksheet (e.g., in Excel) that will perform the required calculations for that day. Your worksheet should do the following, and a copy must be handed in using the mock numbers provided:
- The worksheet should accept 260 nm and 280 nm absorbance readings for RNA samples.
- Mock numbers: 0.524, 0.255 for sample 1; 0.427, 0.212 for sample 2.
- The first calculation should be the 260:280 ratio.
- The next parameter that the user should be able to specify is the RNA dilution factor. Calculate what this is based on the Day 4 protocol.
- If you cannot figure it out, use a value of 200 for now.
- The next calculation should be the RNA concentration in μg/mL. Find the conversion factor for absorbance to RNA concentration in the Day 4 protocol.
- Now calculate the RNA concentration in μM, assuming sample 1 is 6-5 and sample 2 is 8-12 aptamer. (Molecular weights are listed in - you guessed it! - the Day 4 protocol.)
- The user should now be able to input the volume of RNA they have. Use 75 μL for both samples for now.
- Calculate the total nmols of RNA in each sample.
- Calculate how much total volume the RNA should be in to be at a concentration of 8 μM.
- Finally, calculate how much volume needs to be added to the existing RNA to reach that concentration.
- In a separate location on your sheet (or manually), calculate the volume of an 8 μM solution required to have exactly 1.4 nmol of RNA.
- The worksheet should accept 260 nm and 280 nm absorbance readings for RNA samples.
- Prepare a figure depicting your PCR gel results (from Day 2) with an appropriate caption. Also write the portion of your Results (about a paragraph) describing this experiment.
- To ensure that you are making steady progress on the computational assignment, you should begin the MEME analysis (exercise and question 1) and hand in a draft of the associated figure.
- Complete the first portion of the writing exercises from the Day 2 handout. Jargon #2, Readability #2, and Brevity #4 are due on Day 4; the rest of the exercises are due on Day 5.
Reagent List
- G7 buffer (1X, final conc)
- 200 M HEPES-KOH, pH 7.5
- HEPES = 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- 2 mM spermidine
- 40 mM dithiothreitol (DTT)
- 30 mM MgCl2
- 100 μg/mL bovine serum albumin (BSA)
- 200 M HEPES-KOH, pH 7.5
- 1N KOH
- Pyrophosphatase, 100 U/mL in
- 100 mM HEPES-KOH, pH 7.5
- 1 mM DTT
- 50% glycerol
- T7 RNA polymerase
- gene expressed in a pQE30 plasmid (discontinued, originally from Qiagen)
- purified from E. coli
