Flash and JavaScript are required for this feature.
Download the video from iTunes U or the Internet Archive.
Topics covered: Molecular dynamics
Instructor/speaker: Greg Pintilie

Project: Molecular Dynamics
Related Resources
(PDF)
The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To make a donation or view additional materials from hundreds of MIT courses, visit MIT OpenCourseWare at ocw.mit.edu.
PROFESSOR: Greg was not here yesterday. He was away. And so this is work he's largely done on his own again. He's done a molecular dynamics simulator. So he's going to tell us a little bit about that, and then after that we'll do course evaluations, hand out some awards, have some cake, and then after that some pizza, and then play same PlayStation 3s. All yours.
GREG PINTILIE: OK, so I'll talk about the make general molecular dynamics algorithm and then parallelization approaches. So molecular dynamics is based on a potential energy function, which includes two terms, bonded terms and non-bonded terms. And bonded terms are split up into three sub terms. So don't confuse bonded with-- actually, don't think that these are non-bonded. These are actually all sort of grouped as bonded.
I know it's a little bit confusing, but right now I'm dealing just with the sort of bonded terms. And what that means is that it deals with atoms that are connected together covalently. So for example, the bonds term reflects some energy due to a displacement between two atoms, which are certain distance apart. There is an equilibrium distance that they like to stay at, and if they vary from that distance the potential increases as a squared of that separation distance.
The angles term represents, basically, the deviation from an equilibrium angle between three atoms. So if you imagine these three nodes as three atoms, if this angle varies away from a certain equilibrium theta or angle, then the potential energy is also a quadratic term, which tends to bring the three atoms to have a certain equilibrium angle.
And then, finally, the dihedral and improper term is probably one of the more complicated ones. The improper term basically deals with four atoms. So if you imagine this is one, two, three, and four. These three atoms, if you put two planes through them, the angle that those two planes make to each other is given a certain equilibrium angle again.
And this should also be a quadratic term, but the energy also varies quadratically as that angle is deviated from. An actual dihedral is the rotation along a bond between the two central atoms. And this energy term has actually a cosine form. So as you rotate it along this bond, say for example, the energy will very sinusoidally. And there will be a restoring potential that will tend to make it either a line, or sometimes it will make the atom stagger.
So all of these parameters are very sort of molecule and atom dependent. And all of these parameters are available, or have been built over many years by different methods like ab initio calculations or experimental calculations. But basically, if you plug those parameters in, for example, here k represents the spring constant, right? Because they're basically just like spring constraints more or less. Or in this case, it will tell you how far up the potential, like what this value will be at the maximum, and will, for example, in this case, will specify how many fluctuations you have in the energy function.
And the other parameters are usually the theta, the equilibrium theta, or the equilibrium, or the equilibrium bond length. So this is kind of what you can expect for it to look like as molecules will tend to move. The non bonded terms include van-der-Waals terms and electrostatic terms. So van-der-Waals forces occur between any two atoms, and if they're far enough apart, they will attract to each other according to this formula. And if they're too close together, so at equilibrium distance between them the energy is a minimum, but as they get any closer there will be a very strong group repulsive force, which is the r to the 12th term. And as they get further apart, there is an attraction force, which varies as r to the sixth.
And then the electrostatic force is just a simple Coulombic form. So you basically take two atoms that are, say oppositely charged, will attract each other. If they're positively charged they will repel each other. And it's just a matter of multiplying the charges, and then dividing by r to get the energy. OK, so to do molecular dynamics, basically, you have to take derivatives of all those formulas. I just showed you. And I didn't do that all myself. It was pretty hard.
I just got some code from some current existing molecular dynamic simulators. But basically, you take the derivative. And that basically equates to the force, right? The typical Newton Newtonian model. And that gives you the force in every atom, and then using that force and integration scheme, the leapfrog Verlet integration scheme just gives you the velocity at the next half time step. And then using that velocity at the half time step, the positions of each atom is updated based on the positions at the previous time step, and then some value.
So I will show you a little bit of what it looks like on a simple molecule first. So here's a sort of simple molecule, and it has just an initial configuration that is just sort of random. And so one technique that is done is first the geometry is minimized, which means that you just take small steps in the direction of the gradient until you reach the energy minimum. And if I do that, I sort of reach sort of the minimum configuration that this molecule likes to be in.
And then, if I add some thermal noise, which means that I just set the initial velocities of the atoms to some random value based on the temperature, then the molecule starts sort of jiggling around. And the nice thing about the Verlet integration scheme is that it's actually just right, like the energy is conserved in the system, although it fluctuates a little bit. So a lot of integration schemes the energy tends to be either decrease or increase, so the system explodes. But this integration scheme is very stable.
One thing that you saw there is, so I just talked briefly about kinetic energy. This is kind of how the initial velocities are set based on the Boltzmann constant and the temperature. And the other thing that molecular dynamics usually do is called Langevin dynamics. And in this case, some random velocity vector is added to the normal velocity, which just sort of simulates collisions with other molecules in the environment. So this makes the simulation look a bit more realistic. So instead of the molecule just sitting there in space, this random sort of factor adds some movement.
And there's also a damping constant, which sort of opposes the velocity by some constant. So if you didn't add any more sort of randomness into the system, the system would just eventually go to be completely still. So there would be no more movement, because this is kind of like a damping sort of factor. And there are some properties of this random vector that sort of make some physical sense. For example, it's independent of previously picked random vectors, and it also depends on the temperature. So that if you keep adding this random factor to the simulation, the temperature will always stay constant.
Another thing to consider is that these molecules, if you try to think of them as they are in a sort of typical biological system, is that they'll be solvated in water. So in that case the Coulombic term is not actually that simple. You have to include some sort of dielectric constant. So for example, one that is typically used and is pretty simple is that the constant is just the distance. So that makes the electrostatic energy dependent on one over r squared rather than 1 over r. So that means that the electrostatic forces drop off a lot faster as the atoms get further and further apart.
So for example, so two molecules that are oppositely charged, if you just put them in vacuum, they will eventually come close together. But if you add this term, the electrostatic attraction will be much weaker. So it will simulate them being solvated in water. And they will come together eventually but probably a lot slower. Another thing that is typically-- so I'll talk a little bit about the non-bonded forces right now, which are, again, the electrostatic forces and the van-der-Waal's forces.
So generally, what has to be done is, if you consider any single atom like say this one over here, well, so first of all the bonded forces basically include just the atoms that are connected to it, and that's it. So it's usually the fastest step in the computation. But for non-bonded forces, you have to consider basically every other atom in the system. And I didn't include all of the atoms here, but basically just to give an idea, they could be atoms that are really far apart.
So generally, what's done is there is some cut off radius, outside of which those forces aren't considered. And because the forces drop off really fast, sometimes that's considered to be good enough for a lot of simulations. So that's a consideration to take into account when doing these simulations. OK, so here's the basic molecular dynamics algorithm. So you specify how many steps you want to run it to. And each time step is one femtosecond. So very small.
So usually what happens is you can find the atom pairs. So that means all the non-bonded interactions, you have to make a list, basically an n squared list. If you have n atoms, you can expect to have about n squared atom pairs. But if you consider a cut off, then what's usually done is only atom pairs within that cut off are considered and included into that list. And that computation is pretty expensive, d it's sometimes only done only every, say every ten steps.
And atoms don't really move that much. So it's usually pretty accurate. And if you use a cut off of, say 15 angstroms, and then, well, the forces have dropped to almost zero by 12 angstroms, then that's pretty accurate. So this is usually an optimization. It optimizes the simulation a little bit. The next step is to compute the bonded forces. So that includes the bonds, angles, dihedrals and improper angles.
And then compute the non-bonded forces, so that iterates over all the atom pairs. And then the integration is done to obtain the new atom positions. So here's, running on the PlayStation, this is kind of roughly how this is just running on the PPU alone. So finding the atom pairs, if I don't use a cut off, I can just compute the atom pairs list once, and then never do it again. So that's why I put here 0. But otherwise, generally takes a lot of time to compute the atom pairs, because it involves getting the distances between every two atoms.
And this is a pretty big system, maybe about 1,000 atoms. So that's why. So these times are not really representative of smaller systems.
AUDIENCE: What is BF [INAUDIBLE]?
GREG PINTILIE: BF, so that's done using the brute force approach. So that means that you check every two atoms. And this is using a KD tree, so that breaks space up. So then checking the neighboring atoms is n log n. Or it's actually a constant factor after you've built the tree. Building the tree is n log n. So using that kind of an algorithm, you get much better improvement. So this is a scaling of increasing the system size by two in the number of atoms.
So say going from 1,000 to 2,000 atoms. The time too, using a brute force approach, increases exponentially. Whereas, using a KD tree, it actually just about doubles. So that kind of shows that using a better algorithm sometimes for these things is probably much better than trying to parallelize it. Doing the bonded forces, say about 50 milliseconds, and with cut off it's the same time, because it doesn't really affect the step.
But then computing the non-bonded forces is actually the biggest chunk of time. So that takes about 1.4 seconds for the system. And then this is considering a million pairs. With a cut off there is less pairs to consider, but the time is still the dominant time basically. So generally, for molecular dynamic simulations this is the dominant time factor. And that's what people really focus on, trying to speed things up. And then the integration is really fast, because it just loops over the atoms once.
So there is basically three different parallelization approaches that I've come across in the literature. And one of them is referred to as force decomposition. So if you think of a force operation being, say a non-bonded force operation where you consider two atoms, and you compute the force the two, van-der-Waals and electrostatic forces, one easy way to split this up amongst, say six processors, is to just give each processor, say n divided by 6 of these force operations.
So you have to send each processor both atom positions, and then some other properties of the atoms, such as the mass and-- actually not the mass, but the charges and the minimum radius. And basically, once that computation is done, the processor just has to either store or return the force on both atom. And it has to come back to sort of like the main processor where all the forces are added up, and then the integration is done.
So what I just described is sort of the approach I took. And it's sort of an easy approach to sort of think of if you couldn't store, say every atom on every processor. Because you just have to send each processor a certain number of these force computation units. And the processor does then and then returns the forces. And then you can just add up all the forces on every atom and do that. So basically here is the algorithm how it works roughly. And so basically, first set up the SPUs, send the control box with, say the addresses where a these force operations will be stored.
The bonded forces are computed on the PPU, just for simplicity for now. And then the computation for the non-bonded forces, basically, all the non-bonded forces operations remaining are considered. And then they're split up into blocks. So each SPU sent a block with, say a number of force operations. And in this case it was 200. So the control block is sent to the SPU, the SPU is told to start processing.
And then the PPU waits for each SPU to finish. And then once the SPU is finished, the forces are added to the two atoms that are involved in that force computation. And then this loop is done once all the SPUs are finished. And then it goes, in this repeats until there is non-bonded operations remaining. So for example here, just to illustrate this, let's say these are all the non-bonded operations I have to do. And I'm going to use two SPUs. And each SPU can only store, say three force operations.
So each SPU is sent by a block. I mean that, say these three operations are sent to each SPU. The SPU does them. The next block is sent to the next SPU at the same time, and then that SPU also does it. And this is iteration 0 for example. And this is done until all of the force computations are done. So here's a timing. So you might imagine that, as I said, there is there is a lot of communication overhead over here.
So here's how this performs. So on the PPU, the MD simulation per time step, runs in 310 milliseconds. On one SPU, the time was up to 630 milliseconds. So there is a lot of communication overhead here. On two SPUs it goes back down to 390. And as the number of SPUs is increased, it gets lower and lower. But it's not that much better than really running it sequentially. And the reason is that the way I implemented is pretty rough.
One of the limitations, I mean one very obvious optimization that should be done is that SPU basically waits for this whole block to arrive, and then starts computation. But really what I should have done is once one force computation arrives, you can already do that computation. Instead of waiting for all of them to be transferred and then starting to do the operation. So that I think that could've been a lot better.
AUDIENCE: [INAUDIBLE]
GREG PINTILIE: No.
AUDIENCE: [INAUDIBLE]
GREG PINTILIE: No. So right, so this is sort of the simplest approach, I mean sort of a simple approach to implement on this architecture. And it sort of scales well as the system grows. So if you can't store every item, for example, in every SPU, this doesn't really care. The system can grow as large as you want. There's some other approaches that are done sort of in the literature. I didn't implement these, but another one is called atomic decomposition.
So here, if you have, say a number of atoms, you send each one of these atoms to one SPU. And then that SPU is solely responsible for computing the forces on it, and also integrating to get the new positions. And then all you have to communicate is the atomic positions. But this is a little bit more complicated, because then every SPU has to know about all of the force computations that you want to do. So for example, in this case it's just a non-bonded, but the SPU also has to know about--
AUDIENCE: So that would be like the same example that we did in [INAUDIBLE].
GREG PINTILIE: Yeah, it would be. Yeah, the only problem with that approach is that if the system was much larger than the SPU could store, and you couldn't store every single atom on the SPU, then an operation might refer to atoms that are not there. So yeah, it would be similar to that. But it would get more complicated in this case as a system grew, because you'd have to keep track of where each atom is and then implement communication. If you don't have a certain atom, to get to that atom position and use it in the force computation. So yeah, this approach, OK, I'll talk a little bit more about this. This is a--
AUDIENCE: [INAUDIBLE] I mean not SPU, but [INAUDIBLE].
GREG PINTILIE: That's true, yeah. So you only sort of give a certain number of atoms to every SPU, and then, yeah. That's true. And then you would have to sort of store every single force that that atom is included in on the SPU.
AUDIENCE: [INAUDIBLE]
GREG PINTILIE: Yeah, like when this will become a problem is, say the system was much larger than you had local store. Then say you had like 1/10 of the system on SPUs and then 9/10 of the system on the PPU.
AUDIENCE: [INAUDIBLE]
GREG PINTILIE: Yeah, it would be hard to think of it scaling properly. So a special case of this atomic decomposition is actually spatial decomposition. OK, I'm almost done. So a special case of spatial decomposition. So what makes this approach sort of not good, and it sort of involves a lot of communication, is just this fact that some of these force computations, or force operations that you have could include atoms on different units. So if you don't split up these atoms sort of smartly, then you could end up having to communicate a lot while computing these forces.
So spatial decomposition tries to address this. And basically, you store atoms that are close to each other. So say I had the system, and I wanted to split this up on six SPUs, then right, I would give each SPU, say a certain area of space. But you could see right away what the problem with this is is that it's not load balanced. So some SPUs could not have too much work to do. And there's still some communication, because you have to communicate between neighboring SPUs, say some certain cases.
But the communication is still probably a lot better than in the previous case, just for atomic decomposition. So here's the state of the art on parallelization MD. And it's implemented in this program called NAMD, which stands for Not Another Molecular Dynamics simulator. So initially, NAMD 1.0 used solely a spatial decomposition. And they try to address this load balancing by actually creating many more patches than you have processors. And then you just sort of split these patches up, load balance them amongst the different processors.
That didn't really work. So NAMD 1 didn't parallelize very well. So what they added later was they added this idea of patch objects, and also compute objects. So basically a patch object puts a number of patches on a processor, but then you can also create compute patches, which include all of these force operations. And these force operations are also equally distributed amongst processors. And then there's a lot communication between these two.
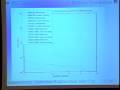
And it started off with sort message oriented communication. Then it moved to a dependency oriented communication so that each compute object would sort of signal which atoms it needs. And it also sort of optimized things by doing communication and force computation at the same time. So it did that really well. And actually it had really good parallel performance. So on this graph it shows the number of processors times the time per time step. And as the number of processors is increased, I mean optimal parallel performance would be a straight horizontal line, which is pretty close to what they achieve.
And they also did it on many thousands of processors on Blue Gene. But as you might expect, the more processors you add, the step time actually still sort of levels off at a certain time. So I guess I'll skip the sort of live demo I was going to do, but I'll just show one more thing. So here is sort of the kind of systems that I was looking at. This is actually a simulation that was done on one of the PPUs and then recorded in positions file. And there is five different molecules here.
So generally, what happens is first they're just placed randomly. Then they're an overlap just very coarsely, to make sure that huge forces are not entered into the system. And I'll just color each one differently. So here's what happens during the simulation. And here, in this simulation, actually, the positions were recorded every 20 time steps. So that's why it sort of seems to go a lot faster. If I did it on a single molecule, I actually done that first.
So here what a single molecule of this looks like. And this is a protein with eight residues, which is pretty big. And this is running in real time on my computer right now. So like on a small system like this, it runs reasonably fast. But as you add more and more of these things, the computations do get pretty expensive. So parallelization really helps. Well, not in what I implemented, but generally. Generally, as you saw, NAMD can do a pretty good job at doing it. Oh OK, I didn't minimize the system well enough. But yeah, that's about it.
AUDIENCE: Great. [INAUDIBLE]
AUDIENCE: So basically, on the SPUs, you managed to get them synchronized properly, data in and data out?
GREG PINTILIE: Yeah, exactly. So that's kind of to the extent that I went. And that still took a long time. So [INAUDIBLE].